research
We study how development and environment interact—that is, by developmental plasticity—to translate genotypes to phenotypes, how this interaction evolves, and how it influences the diversification of form.
The models for our research are nematodes, which lead virtually every lifestyle known to animals and include microbivores, omnivores, predators, and parasites of insects, vertebrates, and plants. The functional diversity of nematodes, which may be the most abundant and species-rich animals on earth, is reflected in their mouthparts. Our lab aims to understand the evolution of this diversity through approaches integrating developmental genetics, phylogenetics, genomics, and natural history.
The main reference point for our research is the shark-tooth nematode Pristionchus pacificus.
This species has a structural innovation, moveable teeth that allow
omnivorous feeding on bacteria, fungi, and even other nematodes.
Predatory feeding by P. pacificus
In response to starvation and crowding, P. pacificus larvae develop into adults of one of two distinct feeding morphs
("eurystomatous" and "stenostomatous"). In this polyphenism (i.e., discrete developmental plasticity), the stenostomatous morph grows rapidly on a diet of bacteria, whereas the eurystomatous morph, which is more complex in form, has higher
fitness than the stenostomatous morph when fed nematode prey (Serobyan et al., 2014).
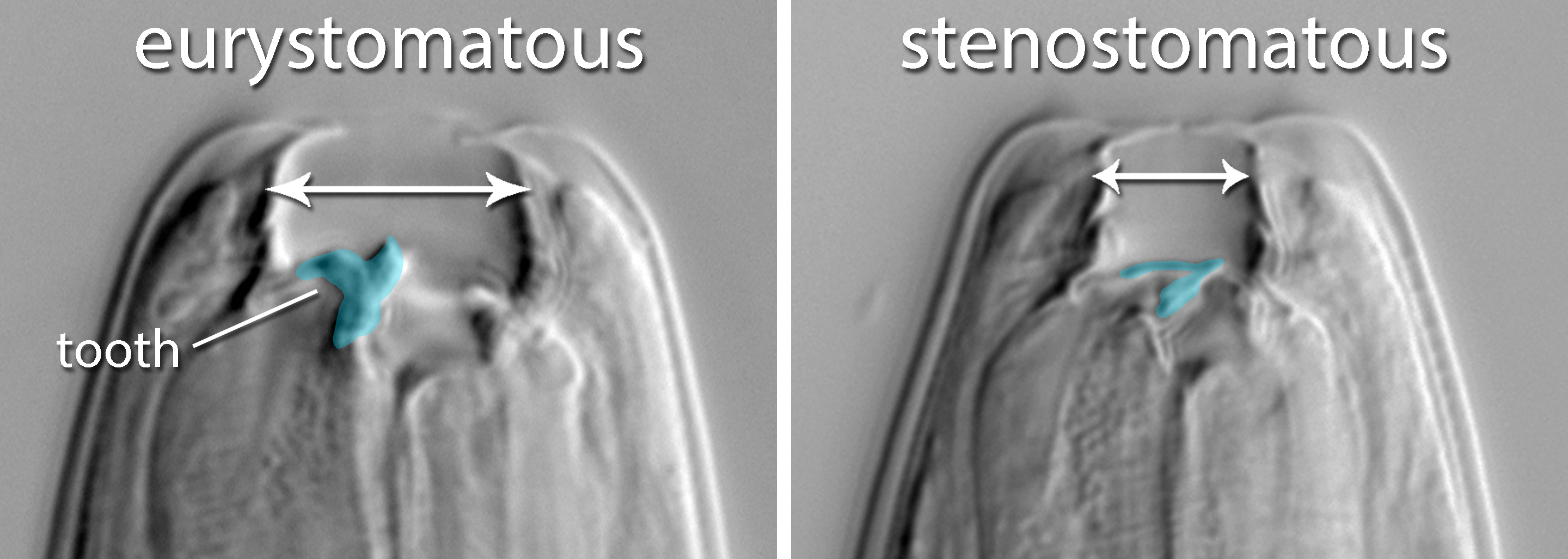
Mouthpart polyphenism in P. pacificus
The ability to make major phenotypic differences from a single genotype, as shown by P. pacificus, may act as a facilitator of novelty and diversity (Susoy et al., 2015).
Research in the lab tests this principle at a molecular level by investigating directly the genes that regulate developmental plasticity and their significance for downstream molecular evolution. Making this research practical is the analytical toolkit available for P. pacificus, a self-fertilizing species with a short (four-day) generation time. Because teeth are what allow these nematodes to be
predators (Nicholson et al., 2024), the interplay of ecology with genetic mechanisms for plasticity can be studied directly in this system.
A few goals of our research are:
1) A molecular understanding of how polyphenism is regulated;
2) To know how plasticity regulators diverge to produce new phenotypes;
3) To determine plasticity's impact on how genes and gene-networks change to produce morphological novelties.
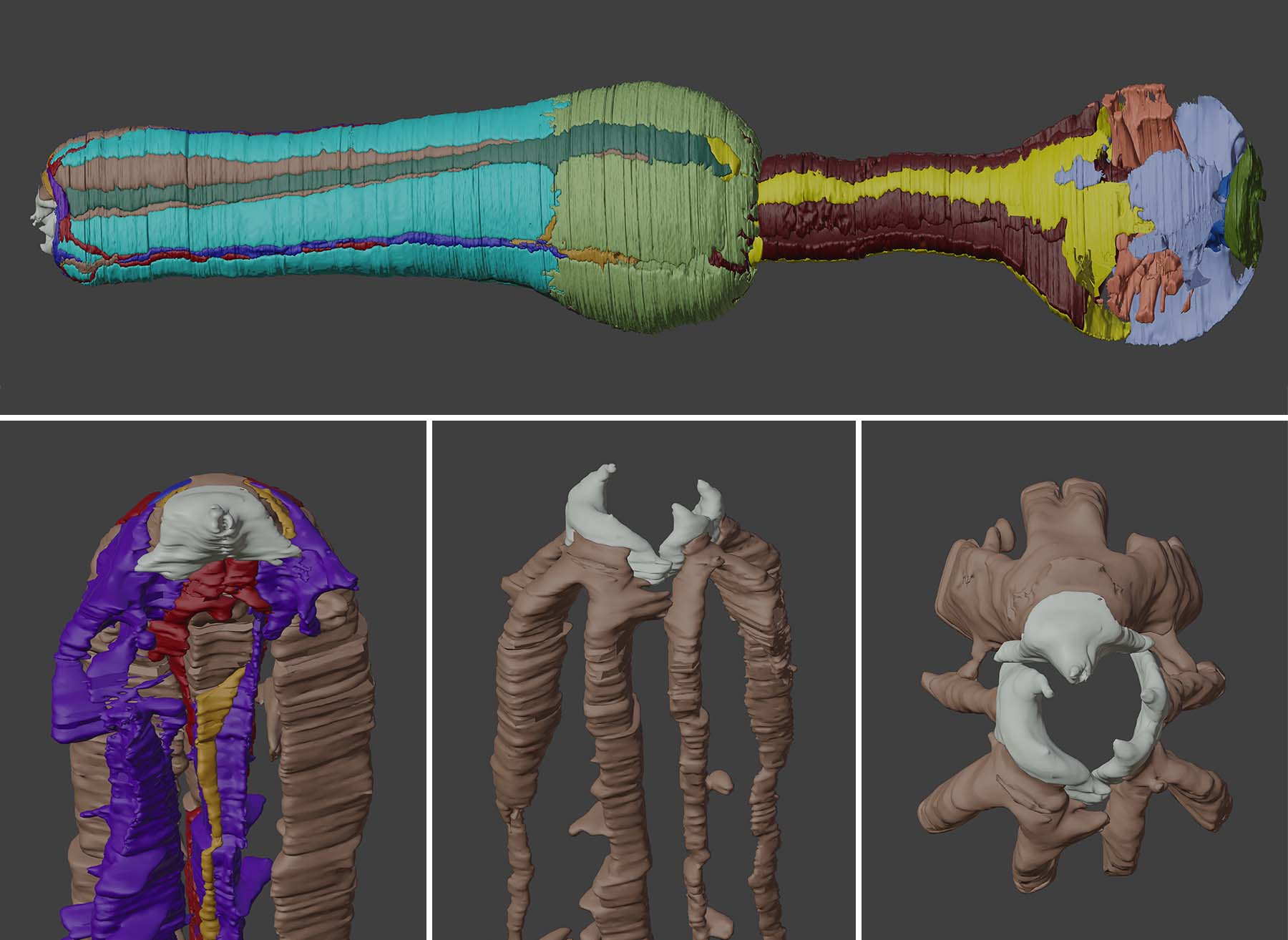
Reconstructed feeding structures of P. pacificus
The moleclular regulation of developmental polyphenism has been accessed through the use of P. pacificus as a model. As a proof of principle, a switch gene—which encodes the sulfatase EUD-1 (eurystomatous-form-defective)—was found to execute a switch for the mouth dimorphism (Ragsdale, Müller et al., 2013). This factor is a gene that has functionally specialized following a series of gene duplications (Ragsdale & Ivers, 2016; Biddle & Ragsdale, 2020).
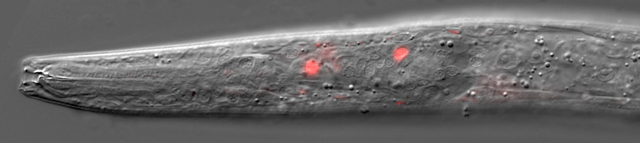
Expression of eud-1 in neurons channeling a polyphenism switch
We have since used eud-1 as a tool to revealing other molecular factors making up the mechanism of the polyphenism switch. Specifially, we have taken an unbiased approach—forward, suppressor screens—to identify the genes involved. One of these genes is another lineage-specific one, which encodes the sulfotransferase SEUD-1 (suppressor-of-eud-1), whose expression is both environmentally influenced and localized to polyphenic tissue (Bui, Ivers & Ragsdale, 2018). Acting downstream of these two enzymes is NHR-40, a nuclear receptor that also fully toggles the switch (Kieninger et al., 2016). Together with NHR-40 and another nuclear receptor, NHR-1, a recent duplicate of the Mediator subunit MED15 canalizes the alternative morphs, thereby achieving polyphenism (Casasa, Katsougia & Ragsdale, 2023).
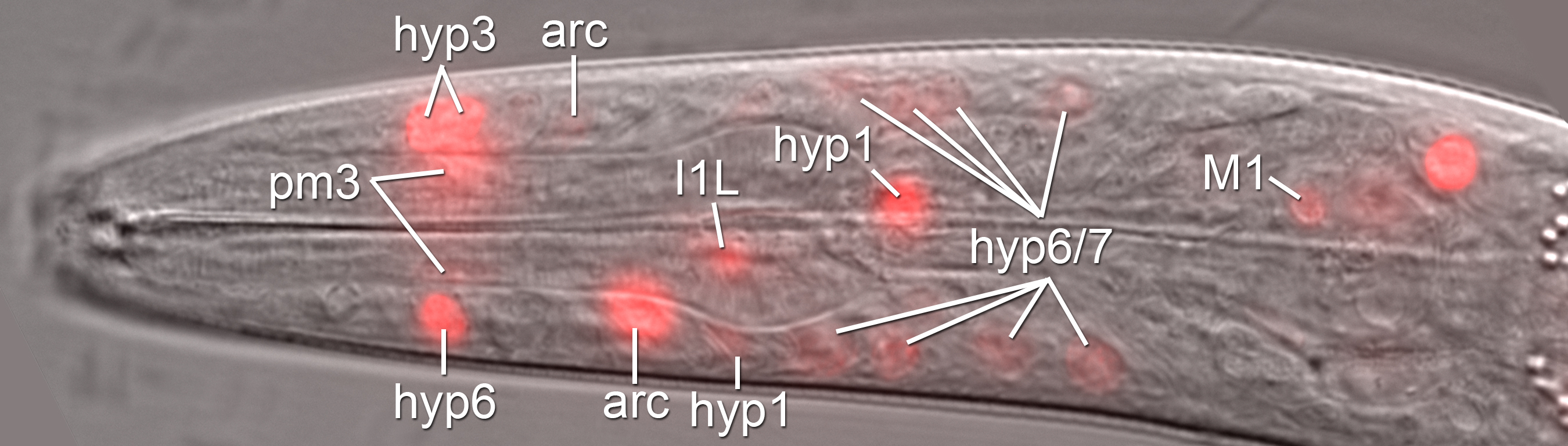
seud-1 expression in cells that produce dimorphic mouthparts
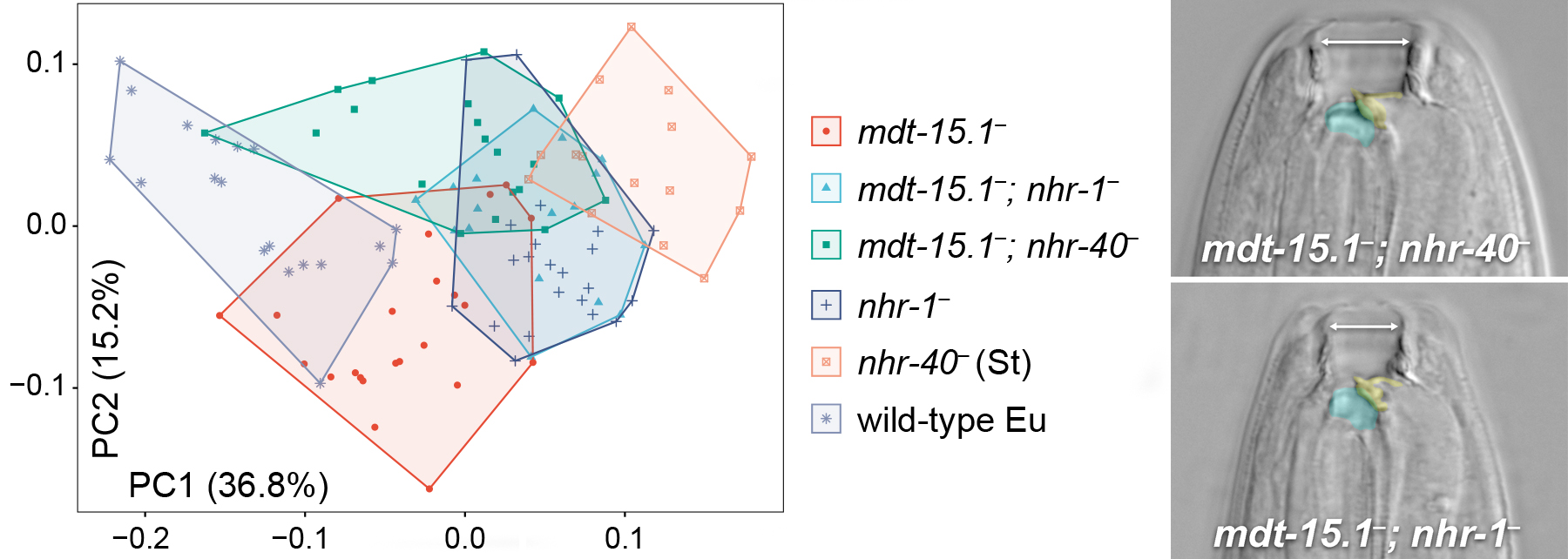
Polyphenism broken by mutants into a continuum of mouth phenotypes (PCA, geometric morphometrics)
Because the switch mediates how morph-specific target genes ("environmentally sensitive loci") are expressed, changes to the switch should mediate how often they are exposed to selection. Therefore, an understanding of the polyphenism switch will enable us to test whether and how changes to its molecular components influence evolution of plastic morphologies. Complemented by a cell-by-cell anatomical reference of the plastic trait (Harry et al., 2022), our findings from forward genetics have begun to reveal this switch's molecular logic.
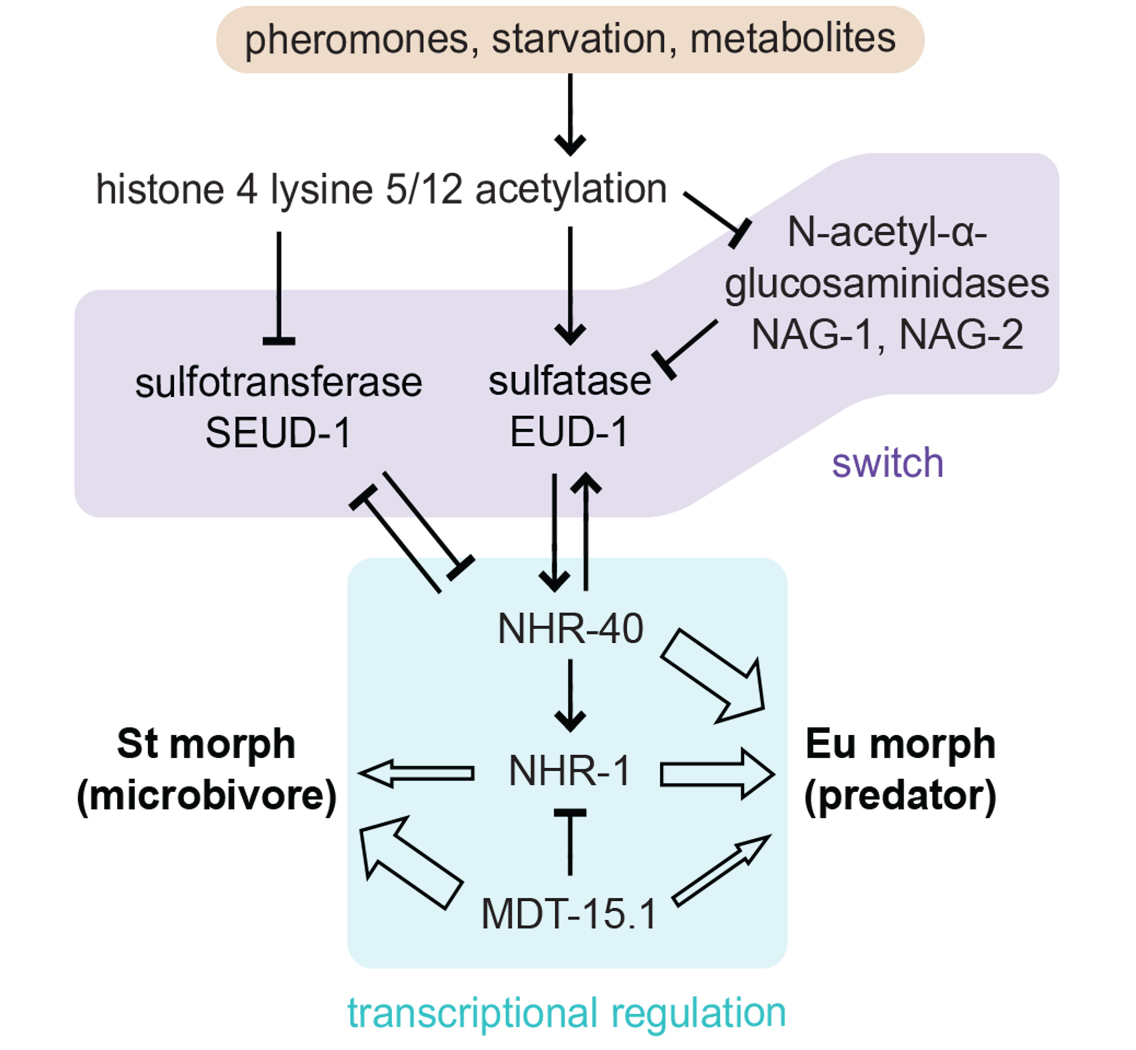
Regulatory model for polyphenism in P. pacificus
Using genetic manipulations in this system, we have also begun to identify the targets of the switch, whose expression indeed underlies phenotypic variation in the polyphenism (Bui & Ragsdale, 2019). This suite of targets has evolved through the network integration of ancient plastic responses (i.e., diapause, metabolism) with highly connected genes undergoing rapid evolutionary turnover (Casasa et al., 2021).
Following these insights, our current aims are to know (i) how the genetic targets of a polyphenism switch instruct alternative morphologies, (ii) what genes' variation explains the divergence of polyphenism, and (iii) the molecular effects of genetic assimilation on originally polyphenic traits. To meet these aims, we are studying polyphenism mechanisms at the scales of both microevolution (natural variation and experimental evolution) and macroevolution (functional genetics in other species).
At the microevolutionary scale, we are using recombinant inbred lines to map quantitative trait loci influencing the polyphenism, and we are leveraging the rapid life-cycle of shark-tooth nematodes to determine how this polyphenism evolves in real time. Together, these approaches will allow us to identify what generalizable genetic features impact plasticity's capacity to change.
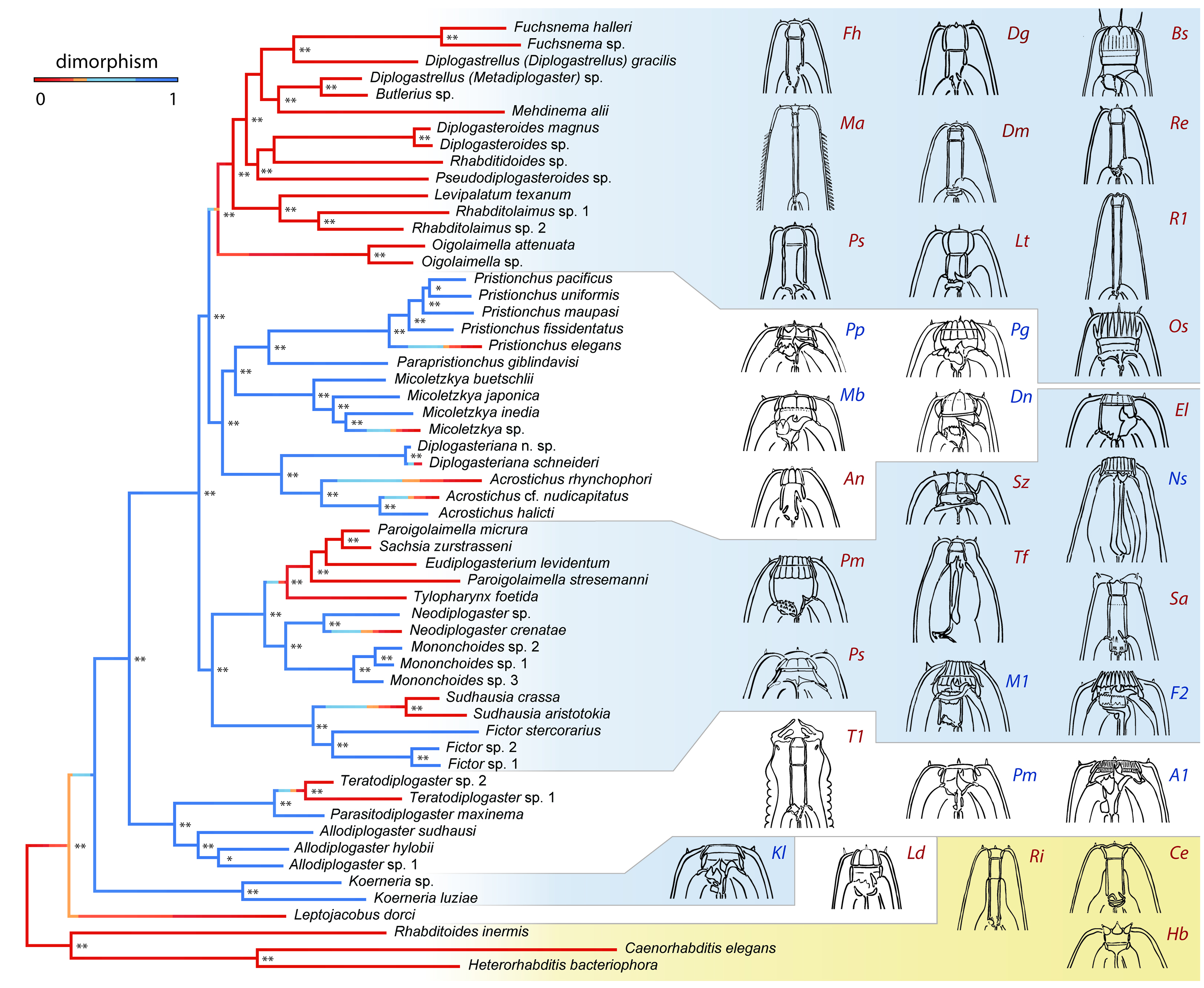
Supporting work at the macroevolutionary scale is a deep source of morphological and life-history variation across the nematode family Diplogastridae, many species of which are easily kept in laboratory culture. Drawing on this resource, we have used reverse genetics in another shark-tooth nematode species (P. fissidentatus) followed by molecular evolutionary inferences across the family to identify another factor in the polyphenism, the H3K4 di/monodemethylase SPR-5/LSD1. Defects in this histone modifier allow selection for heritable differences in the polyphenism even in the absence of genetic variation, suggesting an epigenetic path for plasticity evolution in this system (Levis & Ragsdale, 2023).
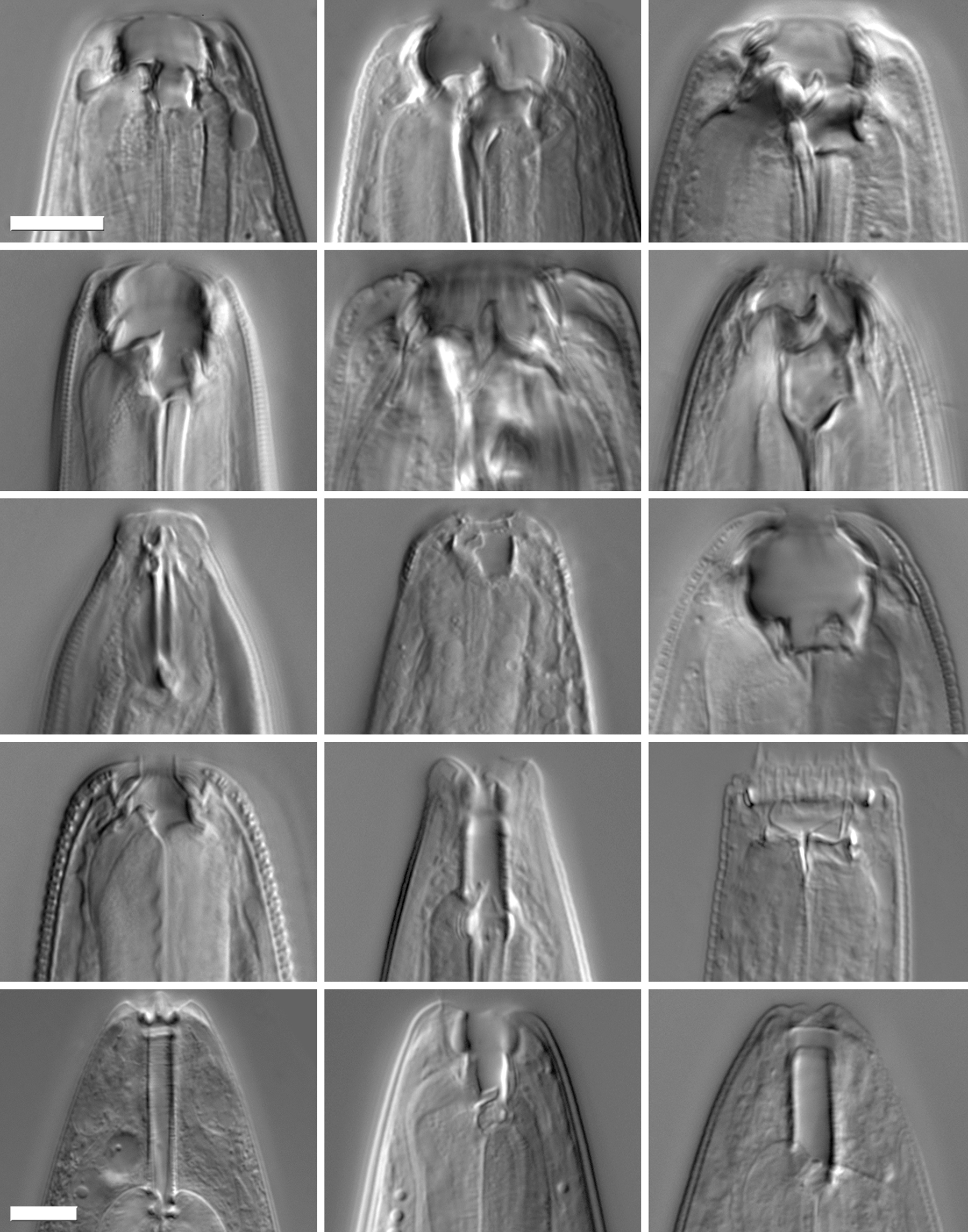
Feeding-structure diversity of Diplogastridae
As we examine the patterns and process of polyphenism evolution, we endeavor to anchor our findings in natural history, so we may know their meaning for evolution in the wild. To this end, we are empirically placing the novel feeding morphologies that characterize Diplogastridae into an ecological context (Ledón-Rettig et al., 2018). In a particularly striking example, polyphenism in one clade of Pristionchus species,
associated with fig wasps in Afrotropical and Australasian figs, has
diverged to include five morphs, each with its own putative ecological
function (Susoy et al., 2016).
Given the diversity of plastic responses, associated morphologies, and
ecological function in Diplogastridae, we can use this system to infer how evolutionary patterns are shaped by the relationship between environment and form.
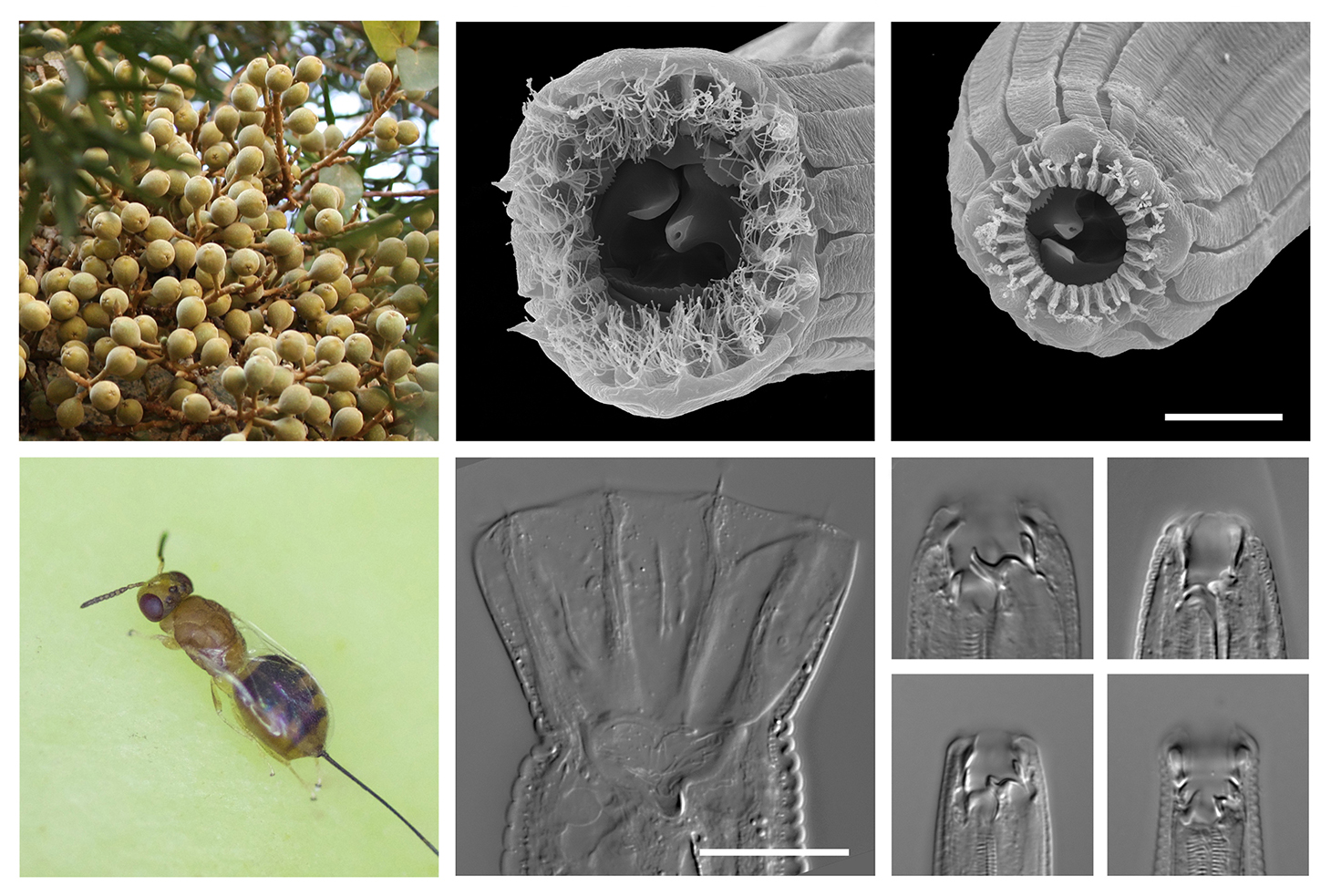
Figs, fig wasps, and the multiple ecomorphs of their Pristionchus associates
Correlation of polyphenism with mouthpart complexity and diversity in macroevolution

